Am I infected with SARS-CoV-2? Is it one of the dangerous variants? Being able to answer these and more questions with a single efficient diagnostic test can be decisive for gauging the spread of disease and selecting the right therapy. In a study published today in the journal "Science", researchers from Würzburg at the Helmholtz Institute for RNA-based Infection Research (HIRI), a site of the Helmholtz Centre for Infection Research (HZI) in cooperation with the Julius Maximilians University (JMU), tackle this challenge with a new CRISPR discovery they translated into a diagnostic platform called LEOPARD.
Most conventional molecular diagnostics usually detect only a single disease-related biomarker. Great examples are the PCR tests currently used to diagnose COVID-19 by detecting a specific sequence from SARS-CoV-2. Such so-called singleplex methods provide reliable results because they are "calibrated" to a single biomarker. However, determining whether a patient is infected with a new SARS-CoV-2 variant or a completely different pathogen requires probing for many different biomarkers at one time.
Scientists from the Helmholtz Institute for RNA-based Infection Research (HIRI) and the Julius Maximilians University (JMU) in Würzburg have now paved the way for a completely new diagnostic platform with LEOPARD. It is a CRISPR-based method that is highly multiplexable, with the potential to detect a variety of disease-related biomarkers in just one test.
How LEOPARD works
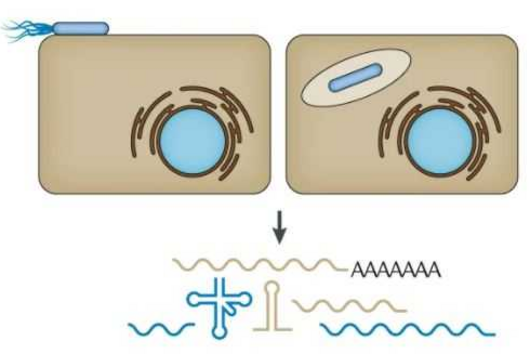
LEOPARD, which stands for “Leveraging Engineered tracrRNAs and On-target DNAs for PArallel RNA Detection,” is based on the finding that DNA cutting by Cas9 could be linked to the presence of a specific ribonucleic acid (RNA). This link allows LEOPARD to detect many RNAs at once, opening opportunities for the simultaneous detection of RNAs from viruses and other pathogens in a patient sample.
The study published today in “Science” was initiated by Chase Beisel, professor at JMU and research group leader at HIRI, and Professor Cynthia Sharma from JMU's Institute of Molecular Infection Biology (IMIB). "With LEOPARD, we succeeded in detecting RNA fragments from nine different viruses,’ says Beisel. “We were also able to differentiate SARS-CoV-2 and one of its variants in a patient sample while confirming that each sample was correctly collected from the patient."
Background
CRISPR-Cas9 is principally known as a biomolecular tool for genome editing. Here, CRISPR-Cas9 function as molecular scissors that cut specific DNA sequences. These same scissors are naturally used by bacteria to cut DNA associated with invading viruses. Whether editing genomes or eliminating viruses, Cas9 cutting is directed by guide RNAs. The guide RNAs found in bacteria must pair with a separate RNA called the tracrRNA. The RNA couple then can work with Cas9 to direct DNA cutting.
An unexpected discovery
The tracrRNA was thought to only pair with guide RNAs coming from the antiviral system. However, the Würzburg scientists discovered that the tracrRNA was pairing with other RNAs, turning them into guide RNAs. Cynthia Sharma, Chair of Molecular Infection Biology II at the IMIB and spokesperson of the Research Center for Infection Diseases (ZINF) at JMU was astounded by this discovery: "When we searched for RNAs binding to Cas9 in our model organism Campylobacter, we surprisingly found that we detected not only guide RNAs, but also other RNA fragments in the cell that looked like guide RNAs. The tracrRNA was pairing with these RNAs, resulting in “non-canonical” guide RNAs that could direct DNA cutting by Cas9."
The LEOPARD diagnostic platform builds on this discovery. "We figured out how to reprogram the tracrRNAs to decide which RNAs become guide RNAs,” says Beisel. “By monitoring a set of matching DNAs, we can determine which RNAs were present in a sample based on which DNAs get cut. As part of the ongoing pandemic, LEOPARD could allow a doctor to figure out whether the patient is infected with SARS-CoV-2, if it’s a unique variant, and whether the sample was correctly taken or needs to be repeated—all in one test.”
In the future, LEOPARD's performance could dwarf even multiplexed PCR tests and other methods. "The technology has the potential to revolutionize medical diagnostics not only for infectious diseases, but also for cancer and rare genetic diseases," says Oliver Kurzai, director of JMU's Institute of Hygiene and Microbiology, which provided patient samples for the study.
Federal and state governments support LEOPARD
“The work highlights the excellent collaborative and interdisciplinary research taking place here in Würzburg,” says Jörg Vogel, director of IMIB and HIRI. "LEOPARD impressively demonstrates that we can cover the full spectrum of complementary cutting-edge research in Würzburg, from the fundamentals of RNA research to clinical applications."
The current study was in part funded by the German Research Foundation (DFG SPP 2141). Funding for the commercialization of LEOPARD is provided through the GO-Bio Initial program from the German Federal Ministry of Education and Research and through a Medical Valley Award from the Bavarian State Ministry of Economic Affairs, Regional Development and Energy.
You can find more information on LEOPARD in this video.
Original article
Non-canonical crRNAs derived from host transcripts enable multiplexable RNA detection by Cas9. Chunlei Jiao, Sahil Sharma†, Gaurav Dugar†, Natalia L. Peeck, Thorsten Bischler, Franziska Wimmer, Yanying Yu, Lars Barquist, Christoph Schoen, Oliver Kurzai, Cynthia M. Sharma*, Chase L. Beisel*. Science, 27.04.2021