
Prof Antoine-Emmanuel Saliba
Single-cell Analysis
Our research and approach
The research group led by Emmanuel Saliba explores host-pathogen interactions in high resolution at the single-cell level. They develop and integrate single-cell genomics, imaging and computational approaches to decipher the microenvironments of individual pathogens and shed light on the heterogeneity of host responses and disease outcomes.
Novel technological leaps are enabling scientists to chart a comprehensive map of the cells across the body, to define their states and to determine their responses to infectious agents in unprecedented detail. Yet how a host either contains the spread of a pathogen, or subsets of pathogens escape host immune surveillance still remains poorly understood.
Emmanuel Saliba’s group investigates RNA and RNA processing as a read-out to determine the cell state of both hosts and pathogens at the single-cell level. Their work, involving pathogens such as Salmonella and respiratory viruses with cell culture models, organoids, and clinical samples, analyses, categorizes and clusters cells to decipher cellular microenvironments and understand infectious disease progression.
They employ single-cell RNA seq, spatial transcriptomics, and RNA imaging to capture the RNA transcript census expressed within a host and pathogen. Further temporal single-cell analysis using RNA metabolic labelling provides insights into the history of a cell. These high-resolution analyses potentially enable the prediction of cell behavior and can unlock gene regulatory networks underlying infectious processes. This work is key in the development of precision diagnostics and therapeutics.
Team members

Prof Antoine-Emmanuel Saliba
Group Leader

Alexander Leipold
PhD Student

Anastasiya Grinko
PhD Student

Ariane Binder
PhD Student

Chia-Ling Chou
PhD Student

Christophe Toussaint
PhD Student

Laura Cecchino
PhD Student

Lisa Chiggiato
PhD Student

Tobias Krammer
PhD Student

Tristan Beste
PhD Student

Dr med Sofie-Katrin Kadel
Clinician Scientist

Nina DiFabion
Technical Assistant
Research projects
Graphical Abstract
Persistent bacterial infections are caused by a minor subpopulation of intracellular pathogens, called ‘persisters’, that reside in different cell types and tissue locations for years asymptomatically. These subpopulations establish a specific cellular organization, which enables them to evade immune surveillance and chemotherapeutic treatment. Histological studies have described complex tissue remodelling during the infection and emerging in vivo studies at the single-cell level have begun to reveal the heterogeneity of infection foci.
However, the cellular architecture of the infection foci and the identification of favoured niches within this complex tissue landscape that impact disease outcome remain open questions. For example, Salmonella are believed to reside in a large variety of cells including macrophages, neutrophils, dendritic cells and epithelial cells. These large cell types exist as a myriad of different sub-classes, which were - until recently - not appreciated. Similarly, within an infected tissue such as the spleen many infected cells escape inflammatory lesions and disseminate into tissues. Therefore, single-cell studies in an in vivo context are necessary to understand the heterogeneity inherent in infected cells, their microenvironment and their function. The Single-cell Analysis group develops and combines in vitro and in vivo single-cell transcriptomics to decipher the cellular organization of infection foci and their functional consequences for infection outcome.
The recent emergence of single-cell genome-wide transcriptomics is proving to be a powerful approach to decipher both cellular identities and function making it possible to study heterogeneity. This is being facilitated by the development of automated platforms that enable the processing of hundreds and thousands of single-cells in parallel. In the context of infection we have pioneered the use of single-cell RNA-seq to investigate heterogeneity in the response of mouse bone marrow-derived macrophages to Salmonella focusing on bacteria with different growth status including non-growing ‘persisters’ that have been linked to recurrent infections. We have described how Salmonella impact the wide spectrum of host polarization and revealed the existence of a subset of macrophages that escape inflammatory and immune activation programs. While providing new insights into the host response, the study was limited to analyzing infected cells from in vitro cultures. The next step is now to decipher the response of single cells of infected tissues, which remains an unmet challenge.
Moreover, the Single-cell Analysis group is fully committed to developing the full potential of single-cell RNA-seq for addressing fundamental scientific questions of infection biology in general, at the HIRI as well as at other locations of the HZI.
In focus
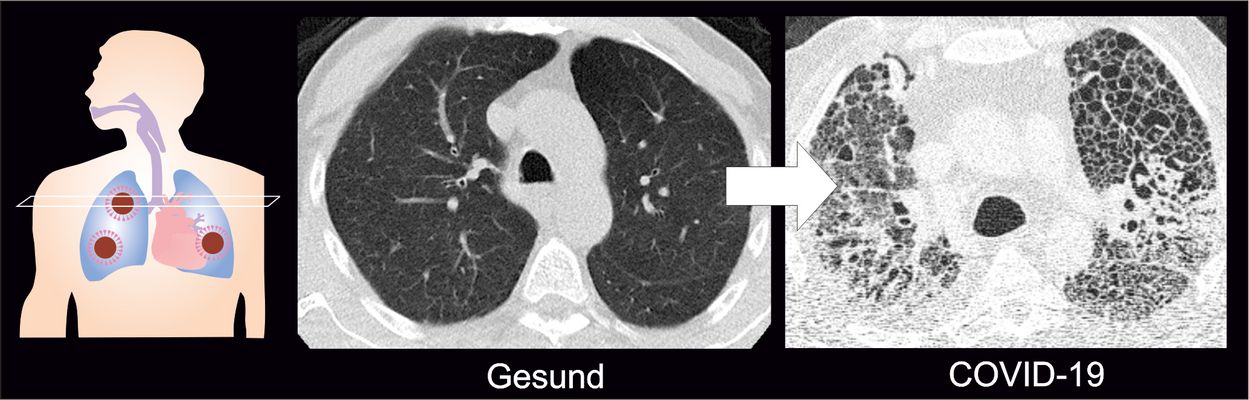
COVID-19-related respiratory failure: Why patients require prolonged ventilation
The majority of patients with severe COVID-19 develop unusually pronounced scarring of the lungs. In a joint study published in Cell, scientists from the Saliba lab and other German research sites report that macrophages — immune cells which engulf and digest foreign substances — play a central role in this regard. Some of the mechanisms involved in COVID-19-related respiratory failure resemble those seen in idiopathic pulmonary fibrosis, a previously incurable disease which causes scarring of the lungs. Misguided wound healing responses resulting in scarring may explain why COVID-19 patients remain in respiratory failure for so long, and why they require prolonged extracorporeal membrane oxygenation (ECMO) support.
Alumni
Oliver Dietrich, PhD Student • Fabian Imdahl, PhD Student • Ehsan Vafadarnejad, PhD Student
Publications
2025
Feature selection methods affect the performance of scRNA-seq data integration and querying
Zappia L, Richter S, Ramírez-Suástegui C, Kfuri-Rubens R, Vornholz L, Wang W, Dietrich O, Frishberg A, Luecken MD, Theis FJ (2025)
Nature Methods (Online ahead of print)
Resolving spatiotemporal dynamics in bacterial multicellular populations: approaches and challenges
Espinoza Miranda SS, Abbaszade G, Hess WR, Drescher K, Saliba AE, Zaburdaev V, Chai L, Dreisewerd K, Grünberger A, Westendorf C, Müller S, Mascher T (2025)
Microbiology and Molecular Biology Reviews 89 (1): e0013824
2024
The life-saving benefit of dexamethasone in severe COVID-19 is linked to a reversal of monocyte dysregulation
Knoll R, Helbig ET, Dahm K, Bolaji O, Hamm F, Dietrich O, van Uelft M, Müller S, Bonaguro L, Schulte-Schrepping J, …, Aschenbrenner AC, Kurth F (2024)
Cell 187 (16): 4318-4335.e20
L-Wnk1 Deletion in Smooth Muscle Cells Causes Aortitis and Inflammatory Shift
Quelquejay H, Al-Rifai R, Silvestro M, Vandestienne M, Ferreira I, Mirault T, Henrion D, Zhong X, Santos-Zas I, Goudot G, …, Jeunemaitre X, Ait-Oufella H (2024)
Circulation Research 135 (4): 488-502
Decoding spatiotemporal transcriptional dynamics and epithelial fibroblast crosstalk during gastroesophageal junction development through single cell analysis
Kumar N, Prakash PG, Wentland C, Kurian SM, Jethva G, Brinkmann V, Mollenkopf HJ, Krammer T, Toussaint C, Saliba AE, …, Gurumurthy RK, Chumduri C (2024)
Nature Communications 15 (1): 3064
Human cytomegalovirus exploits STING signaling and counteracts IFN/ISG induction to facilitate infection of dendritic cells
Costa B, Becker J, Krammer T, Mulenge F, Durán V, Pavlou A, Gern OL, Chu X, Li Y, Cicin-Šain L, …, Erhard F, Kalinke U (2024)
Nature Communications 15 (1): 1745
Circulating NK cells establish tissue residency upon acute infection of skin and mediate accelerated effector responses to secondary infection
Torcellan T, Friedrich C, Doucet-Ladevèze R, Ossner T, Solé VV, Riedmann S, Ugur M, Imdahl F, Rosshart SP, Arnold SJ, …, Kastenmüller W, Gasteiger G (2024)
Immunity 57 (1): 124-140.e7
Sequential Antigen-loss and Branching Evolution in Lymphoma after CD19- and CD20-Targeted T-cell Redirecting Therapy
Duell J, Leipold AM, Appenzeller S, Fuhr V, Rauert-Wunderlich H, Da Vià MC, Dietrich O, Toussaint C, Imdahl F, Eisele F, …, Saliba AE, Rasche L (2024)
Blood 143 (8): 685-696
Alternating high-fat diet enhances atherosclerosis by neutrophil reprogramming
Lavillegrand JR, Al-Rifai R, Thietart S, Guyon T, Vandestienne M, Cohen R, Duval V, Zhong X, Yen D, Ozturk M, …, Riksen NP, Ait-Oufella H (2024)
Nature 634 (8033): 447-456
A novel in vitro tubular model to recapitulate features of distal airways: The bronchioid
Maurat E, Raasch K, Leipold AM, Henrot P, Zysman M, Prevel R, Trian T, Krammer T, Bergeron V, Thumerel M, …, Recher G, Dupin I (2024)
The European Respiratory journal 64 (4)
Neural network-assisted humanisation of COVID-19 hamster transcriptomic data reveals matching severity states in human disease
Friedrich VD, Pennitz P, Wyler E, Adler JM, Postmus D, Müller K, Teixeira Alves LG, Prigann J, Pott F, Vladimirova D, …, Kirsten H, Nouailles G (2024)
EBioMedicine 108: 105312
Characterization and implementation of the MarathonRT template-switching reaction to expand the capabilities of RNA-Seq
Guo LT, Grinko A, Olson S, Leipold A, Graveley B, Saliba AE, Pyle AM (2024)
RNA 30 (11): 1495–1512
Spatial transcriptomics reveals profound subclonal heterogeneity and T-cell dysfunction in extramedullary myeloma
John M, Helal M, Duell J, Mattavelli G, Stanojkovska E, Afrin N, Leipold AM, Steinhardt MJ, Zhou X, Žihala D, …, Riedel A, Rasche L (2024)
Blood 144 (20): 2121-2135
CD8+ T Cells Drive Plaque Smooth Muscle Cell Dedifferentiation in Experimental Atherosclerosis
Schäfer S, Gogiraju R, Rösch M, Kerstan Y, Beck L, Garbisch J, Saliba AE, Gisterå A, Hermanns HM, Boon L, …, Cochain C, Zernecke A (2024)
Arteriosclerosis Thrombosis, and Vascular biology 44 (8): 1852-1872
Characterization of a Human Respiratory Mucosa Model to Study Odorant Metabolism
Mérignac-Lacombe J, Kornbausch N, Sivarajan R, Boichot V, Berg K, Oberwinkler H, Saliba AE, Loos HM, Ehret Kasemo T, Scherzad A, …, Heydel JM, Steinke M (2024)
Journal of Agricultural and Food Chemistry 72 (22): 12696-12706
TREM2 protects from atherosclerosis by limiting necrotic core formation
Piollet M, Porsch F, Rizzo G, Kapser F, Schulz DJ, Kiss MG, Schlepckow K, Morenas-Rodriguez E, Sen MO, Gropper J, …, Binder CJ, Cochain C (2024)
Nature Cardiovascular Research 3 (3): 269-282
2023
Brain-to-gut trafficking of alpha-synuclein by CD11c+ cells in a mouse model of Parkinson's disease
McFleder RL, Makhotkina A, Groh J, Keber U, Imdahl F, Peña Mosca J, Peteranderl A, Wu J, Tabuchi S, Hoffmann J, …, Volkmann J, Ip CW (2023)
Nature Communications 14 (1): 7529
Vector-borne Trypanosoma brucei parasites develop in artificial human skin and persist as skin tissue forms
Reuter C, Hauf L, Imdahl F, Sen R, Vafadarnejad E, Fey P, Finger T, Jones NG, Walles H, Barquist L, …, Groeber-Becker F, Engstler M (2023)
Nature Communications 14 (1): 7660
Microglia-mediated demyelination protects against CD8+ T cell-driven axon degeneration in mice carrying PLP defects
Groh J, Abdelwahab T, Kattimani Y, Hörner M, Loserth S, Gudi V, Adalbert R, Imdahl F, Saliba AE, Coleman M, …, Simons M, Martini R (2023)
Nature Communications 14 (1): 6911
Th17.1 cell driven sarcoidosis-like inflammation after anti-BCMA CAR T cells in multiple myeloma
Leipold AM, Werner RA, Düll J, Jung P, John M, Stanojkovska E, Zhou X, Hornburger H, Ruckdeschel A, Dietrich O, …, Saliba AE, Rasche L (2023)
Leukemia 37 (3): 650-658
Myocardial Milieu Favors Local Differentiation of Regulatory T Cells
Delgobo M, Weiß E, ElDin Ashour D, Richter L, Popiolkowski L, Arampatzi P, Stangl V, Arias-Loza P, Mariotti-Ferrandiz E, Rainer PP, …, Frantz S, Campos Ramos G (2023)
Circulation Research 132 (5): 565-582
Integrated single-cell analysis based classification of vascular mononuclear phagocytes in mouse and human atherosclerosis
Zernecke A, Erhard F, Weinberger T, Schulz C, Ley K, Saliba AE, Cochain C (2023)
Cardiovascular Research 119 (8): 1676-1689
Dynamics of monocyte-derived macrophage diversity in experimental myocardial infarction
Rizzo G, Gropper J, Piollet M, Vafadarnejad E, Rizakou A, Bandi SR, Arampatzi P, Krammer T, DiFabion N, Dietrich O, …, Saliba AE, Cochain C (2023)
Cardiovascular Research 119 (3): 772-785
Rapid neutrophil mobilisation by VCAM-1+ endothelial extracellular vesicles
Akbar N, Braithwaite AT, Corr EM, Koelwyn GJ, van Solingen C, Cochain C, Saliba AE, Corbin A, Pezzolla D, Møller Jørgensen M, …, Melling GE, Shanmuganathan M (2023)
Cardiovascular Research 119 (1): 236-251
Genetic inhibition of CARD9 accelerates the development of atherosclerosis in mice through CD36 dependent-defective autophagy
Zhang Y, Vandestienne M, Lavillegrand JR, Joffre J, Santos-Zas I, Lavelle A, Zhong X, Le Goff W, Guérin M, Al-Rifai R, …, Sokol H, Ait-Oufella H (2023)
Nature Communications 14 (1): 4622
Short-range interactions between fibrocytes and CD8+ T cells in COPD bronchial inflammatory response
Eyraud E, Maurat E, Sac-Epée JM, Henrot P, Zysman M, Esteves P, Trian T, Dupuy JW, Leipold A, Saliba AE, …, Berger P, Dupin I (2023)
eLife 12: RP85875
Lymph node medulla regulates the spatiotemporal unfolding of resident dendritic cell networks
Ugur M, Labios RJ, Fenton C, Knöpper K, Jobin K, Imdahl F, Golda G, Hoh K, Grafen A, Kaisho T, …, Bajénoff M, Kastenmüller W (2023)
Immunity 58 (8): 1778-1793.e10
Cytotoxic CNS-associated T cells drive axon degeneration by targeting perturbed oligodendrocytes in PLP1 mutant mice
Abdelwahab T, Stadler D, Knöpper K, Arampatzi P, Saliba AE, Kastenmüller W, Martini R, Groh J (2023)
iScience 26 (5): 106698
An interferon gamma response signature links myocardial aging and immunosenescence
Ashour D, Rebs S, Arampatzi P, Saliba AE, Dudek J, Schulz R, Hofmann U, Frantz S, Cochain C, Streckfuß-Bömeke K, Ramos GC (2023)
Cardiovascular Research 119 (14): 2458-2468
Advancing massive transcriptional profiling of single bacteria
Saliba AE (2023)
Cell Reports Methods 3 (2): 100416
A primary cell-based in vitro model of the human small intestine reveals host olfactomedin 4 induction in response to Salmonella Typhimurium infection
Däullary T, Imdahl F, Dietrich O, Hepp L, Krammer T, Fey C, Neuhaus W, Metzger M, Vogel J, Westermann AJ, Saliba AE, Zdzieblo D (2023)
Gut Microbes 15 (1): 2186109
Altered and allele-specific open chromatin landscape reveals epigenetic and genetic regulators of innate immunity in COVID-19
Zhang B, Zhang Z, Koeken VACM, Kumar S, Aillaud M, Tsay HC, Liu Z, Kraft ARM, Soon CF, Odak I, …, Schulte LN, Li Y (2023)
Cell Genomics 3 (2): 100232
A MATQ-seq-Based Protocol for Single-Cell RNA-seq in Bacteria
Homberger C, Saliba AE, Vogel J (2023)
Methods in Molecular Biology 2584: 105-121
2022
JAK2V617F mutation drives vascular resident macrophages toward a pathogenic phenotype and promotes dissecting aortic aneurysm
Al-Rifai R, Vandestienne M, Lavillegrand JR, Mirault T, Cornebise J, Poisson J, Laurans L, Esposito B, James C, Mansier O, …, Rautou PE, Ait-Oufella H (2022)
Nature Communications 13 (1): 6592
Postnatal expansion of mesenteric lymph node stromal cells towards reticular and CD34+ stromal cell subsets
Pezoldt J, Wiechers C, Zou M, Litovchenko M, Biocanin M, Beckstette M, Sitnik K, Palatella M, van Mierlo G, Chen W, …, Deplancke B, Huehn J (2022)
Nature Communications 13 (1): 7227
Helicobacter pylori shows tropism to gastric differentiated pit cells dependent on urea chemotaxis
Aguilar C, Pauzuolis M, Pompaiah M, Vafadarnejad E, Arampatzi P, Fischer M, Narres D, Neyazi M, Kayisoglu Ö, Sell T, …, Saliba AE, Bartfeld S (2022)
Nature Communications 13 (1): 5878
Fibroblastic reticular cells mitigate acute graft-versus-host disease via MHCII-dependent maintenance of regulatory T cells
Shaikh H, Pezoldt J, Mokhtari Z, Gamboa Vargas J, Le DD, Peña Mosca J, Arellano-Viera E, Kern MA, Graf C, Beyersdorf N, …, Huehn J, Beilhack A (2022)
JCI Insight 7 (22): e154250
Landscape and age dynamics of immune cells in the Egyptian rousette bat
Friedrichs V, Toussaint C, Schäfer A, Rissmann M, Dietrich O, Mettenleiter TC, Pei G, Balkema-Buschmann A, Saliba AE, Dorhoi A (2022)
Cell Reports 40 (10): 111305
Nonproductive exposure of PBMCs to SARS-CoV-2 induces cell-intrinsic innate immune responses
Kazmierski J, Friedmann K, Postmus D, Emanuel J, Fischer C, Jansen J, Richter A, Bosquillon de Jarcy L, Schüler C, Sohn M, …, Niemeyer D, Goffinet C (2022)
Molecular Systems Biology 18 (8): e10961
Lymphatic migration of unconventional T cells promotes site-specific immunity in distinct lymph nodes
Ataide MA, Knöpper K, Cruz de Casas P, Ugur M, Eickhoff S, Zou M, Shaikh H, Trivedi A, Grafen A, Yang T, …, Gasteiger G, Kastenmüller W (2022)
Immunity 55 (10): 1813–1828.e9
Complement activation induces excessive T cell cytotoxicity in severe COVID-19
Georg P, Astaburuaga-García R, Bonaguro L, Brumhard S, Michalick L, Lippert LJ, Kostevc T, Gäbel C, Schneider M, Streitz M, …, Sawitzki B, PA-COVID-19 Study Group (2022)
Cell 185 (3): 493-512.e25
Interleukin-23 receptor expressing γδ T cells locally promote early atherosclerotic lesion formation and plaque necrosis in mice
Gil-Pulido J, Amézaga N, Jorgacevic I, Manthey HD, Rösch M, Brand T, Cidlinsky P, Schäfer S, Beilhack A, Saliba AE, …, Cochain C, Zernecke A (2022)
Cardiovascular Research 118 (14): 2932-2945
Time-resolved single-cell RNA-seq using metabolic RNA labelling
Erhard F, Saliba AE, Lusser A (2022)
Nature Reviews Methods Primers 2 (1): 331
Type 1 conventional dendritic cells maintain and guide the differentiation of precursors of exhausted T cells in distinct cellular niches
Dähling S, Mansilla AM, Knöpper K, Grafen A, Utzschneider DT, Ugur M, Whitney PG, Bachem A, Arampatzi P, Imdahl F, …, Bedoui S, Kastenmüller W (2022)
Immunity 55 (4): 656-670.e8
2021
Effector differentiation downstream of lineage commitment in ILC1s is driven by Hobit across tissues
Friedrich C, Taggenbrock RLRE, Doucet-Ladevèze R, Golda G, Moenius R, Arampatzi P, Kragten NAM, Kreymborg K, Gomez de Agüero M, Kastenmüller W, …, van Gisbergen KPJM, Gasteiger G (2021)
Nature Immunology 22 (10): 1256-1267
SARS-CoV-2 infection triggers profibrotic macrophage responses and lung fibrosis
Wendisch D, Dietrich O, Mari T, von Stillfried S, Ibarra IL, Mittermaier M, Mache C, Chua RL, Knoll R, Timm S, …, Saliba AE, Sander LE (2021)
Cell 184 (26): 6243-6261.e27
Homozygous BCMA gene deletion in response to anti-BCMA CAR T cells in a patient with multiple myeloma
Da Vià MC, Dietrich O, Truger M, Arampatzi P, Duell J, Heidemeier A, Zhou X, Danhof S, Kraus S, Chatterjee M, …, Saliba AE, Rasche L (2021)
Nature Medicine 27 (4): 616-619
Opposing Wnt signals regulate cervical squamocolumnar homeostasis and emergence of metaplasia
Chumduri C, Gurumurthy RK, Berger H, Dietrich O, Kumar N, Koster S, Brinkmann V, Hoffmann K, Drabkina M, Arampatzi P, …, Saliba AE, Meyer TF (2021)
Nature Cell Biology 23 (2): 184–197
Dysregulated Immunometabolism Is Associated with the Generation of Myeloid-Derived Suppressor Cells in Staphylococcus aureus Chronic Infection
Dietrich O, Heinz A, Goldmann O, Geffers R, Beineke A, Hiller K, Saliba AE, Medina E (2021)
Journal of Innate Immunity: 1-18
The healing myocardium mobilises a distinct B-cell subset through a CXCL13-CXCR5-dependent mechanism
Heinrichs M, Ashour D, Siegel J, Büchner L, Wedekind G, Heinze KG, Arampatzi P, Saliba AE, Cochain C, Hofmann U, Frantz S, Campos Ramos G (2021)
Cardiovascular Research 117 (13): 2664-2676
Time-Resolved scRNA-Seq Tracks the Adaptation of a Sensitive MCL Cell Line to Ibrutinib Treatment
Fuhr V, Vafadarnejad E, Dietrich O, Arampatzi P, Riedel A, Saliba AE, Rosenwald A, Rauert-Wunderlich H (2021)
International Journal of Molecular Sciences 22 (5): 2276
Initial HCV infection of adult hepatocytes triggers a temporally structured transcriptional program containing diverse pro- and anti-viral elements
Tegtmeyer B, Vieyres G, Todt D, Lauber C, Ginkel C, Engelmann M, Herrmann M, Pfaller CK, Vondran FW, Broering R, …, Pietschmann T, Brown RJ (2021)
Journal of Virology 95 (10)
A genome-wide transcriptomic analysis of embryos fathered by obese males in a murine model of diet-induced obesity
Bernhardt L, Dittrich M, El-Merahbi R, Saliba AE, Müller T, Sumara G, Vogel J, Nichols-Burns S, Mitchell M, Haaf T, El Hajj N (2021)
Scientific Reports 11: 1979
2020
Longitudinal Multi-omics Analyses Identify Responses of Megakaryocytes, Erythroid Cells, and Plasmablasts as Hallmarks of Severe COVID-19
Bernardes JP, Mishra N, Tran F, Bahmer T, Best L, Blase JI, Bordoni D, Franzenburg J, Geisen U, Josephs-Spaulding J, …, Schultze JL, Rosenstiel P (2020)
Immunity 53 (6): 1296-1314.e9
Dynamics of Cardiac Neutrophil Diversity in Murine Myocardial Infarction
Vafadarnejad E, Rizzo G, Krampert L, Arampatzi P, Arias-Loza A, Nazzal Y, Rizakou A, Knochenhauer T, Bandi SR, Nugroho VA, …, Saliba AE, Cochain C (2020)
Circulation Research 127 (9): e232-249
LifeTime and improving European healthcare through cell-based interceptive medicine
Rajewsky N, Almouzni G, Gorski SA, Aerts S, Amit I, Bertero MG, Bock C, Bredenoord AL, Cavalli G, Chiocca S, …, Vidal M, Voet T (2020)
Nature 587 (7834): 377-386
Single-cell RNA-sequencing reports growth-condition-specific global transcriptomes of individual bacteria
Imdahl F, Vafadarnejad E, Homberger C, Saliba AE, Vogel J (2020)
Nature Microbiology 5 (10): 1202–1206
Severe COVID-19 is marked by a dysregulated myeloid cell compartment
Schulte-Schrepping J, Reusch N, Paclik D, Baßler K, Schlickeiser S, Zhang B, Krämer B, Krammer T, Brumhard S, Bonaguro L, …, Saliba AE, Sander LE (2020)
Cell 182 (6): 1419-1440
Eleven grand challenges in single-cell data science
Lähnemann D, Köster J, Szczurek E, McCarthy DJ, Hicks SC, Robinson MD, Vallejos CA, Campbell KR, Beerenwinkel N, Mahfouz A, …, Shah SP, Schönhuth A (2020)
Genome Biology 21: 31
Advances and challenges in single-cell RNA-seq of microbial communities
Imdahl F, Saliba A (2020)
Current Opinion in Microbiology 57: 102-110
Tracheal brush cells release acetylcholine in response to bitter tastants for paracrine and autocrine signaling
Hollenhorst MI, Jurastow I, Nandigama R, Appenzeller S, Li L, Vogel J, Wiederhold S, Althaus M, Empting M, Altmüller J, …, Saliba AE, Krasteva-Christ G (2020)
The FASEB Journal 34 (1): 316-332
2019
scSLAM-seq reveals core features of transcription dynamics in single cells
Erhard F, Baptista MA, Krammer T, Hennig T, Lange M, Arampatzi P, Jürges CS, Theis FJ, Saliba AE, Dölken L (2019)
Nature 571 (7765): 419-423
2018
Salmonella persisters undermine host immune defenses during antibiotic treatment
Stapels DA, Hill PW, Westermann AJ, Fisher RA, Thurston TL, Saliba AE, Blommestein I, Vogel J, Helaine S (2018)
Science 362 (6419): 1156-1160
Neonatally imprinted stromal cell subsets induce tolerogenic dendritic cells in mesenteric lymph nodes
Pezoldt J, Pasztoi M, Zou M, Wiechers C, Beckstette M, Thierry GR, Vafadarnejad E, Floess S, Arampatzi P, Buettner M, …, Saliba AE, Huehn J (2018)
Nature Communications 9: 3903
Single-Cell RNA-Seq Reveals the Transcriptional Landscape and Heterogeneity of Aortic Macrophages in Murine Atherosclerosis
Cochain C, Vafadarnejad E, Arampatzi P, Pelisek J, Winkels H, Ley K, Wolf D, Saliba AE, Zernecke A (2018)
Circulation Research 122 (12): 1661-1674
Atlas of the Immune Cell Repertoire in Mouse Atherosclerosis Defined by Single-Cell RNA-Sequencing and Mass Cytometry
Winkels H, Ehinger E, Vassallo M, Buscher K, Dinh HQ, Kobiyama K, Hamers AA, Cochain C, Vafadarnejad E, Saliba AE, …, Ley K, Wolf D (2018)
Circulation Research 122 (12): 1675-1688
Genome organization and DNA accessibility control antigenic variation in trypanosomes
Müller LS, Cosentino RO, Förstner KU, Guizetti J, Wedel C, Kaplan N, Janzen CJ, Arampatzi P, Vogel J, Steinbiss S, …, Sebra RP, Siegel TN (2018)
Nature 563 (7729): 121-125
Tolerogenic Transcriptional Signatures of Steady-State and Pathogen-Induced Dendritic Cells
Vendelova E, Ashour D, Blank P, Erhard F, Saliba AE, Kalinke U, Lutz MB (2018)
Frontiers in Immunology 9: 333
2017
Einzelzell-RNA-Sequenzierung beleuchtet den Infektionsprozess
Saliba AE, Westermann AJ, Vogel J (2017)
BIOspektrum 23 (5): 525-528
New RNA-seq approaches for the study of bacterial pathogens
Saliba AE, Santos SC, Vogel J (2017)
Current Opinion in Microbiology 35: 78-87
2016
A protocol for the systematic and quantitative measurement of protein-lipid interactions using the liposome-microarray-based assay
Saliba AE, Vonkova I, Deghou S, Ceschia S, Tischer C, Kugler KG, Bork P, Ellenberg J, Gavin A (2016)
Nature Protocols 11 (6): 1021-38
Single-cell RNA-seq ties macrophage polarization to growth rate of intracellular Salmonella
Saliba AE, Li L, Westermann AJ, Appenzeller S, Stapels DA, Schulte LN, Helaine S, Vogel J (2016)
Nature Microbiology 2: 16206
2015
The systematic analysis of protein-lipid interactions comes of age
Saliba AE, Vonkova I, Gavin A (2015)
Nature Reviews Molecular Cell Biology 16 (12): 753-61
Lipid Cooperativity as a General Membrane-Recruitment Principle for PH Domains
Vonkova I, Saliba AE, Deghou S, Anand K, Ceschia S, Doerks T, Galih A, Kugler KG, Maeda K, Rybin V, …, Bork P, Gavin A (2015)
Cell Reports 12 (9): 1519-30
2014
A quantitative liposome microarray to systematically characterize protein-lipid interactions
Saliba AE, Vonkova I, Ceschia S, Findlay GM, Maeda K, Tischer C, Deghou S, van Noort V, Bork P, Pawson T, Ellenberg J, Gavin A (2014)
Nature Methods 11 (1): 47-50
Single-cell RNA-seq: advances and future challenges
Saliba AE, Westermann AJ, Gorski SA, Vogel J (2014)
Nucleic Acids Research 42 (14): 8845-60
2010
Microfluidic sorting and multimodal typing of cancer cells in self-assembled magnetic arrays
Saliba AE, Saias L, Psychari E, Minc N, Simon D, Bidard F, Mathiot C, Pierga J, Fraisier V, Salamero J, …, Malaquin L, Viovy J (2010)
PNAS 107 (33): 14524-9
2009
Cellules tumorales circulantes et cancer du sein : méthodes de détection et résultats cliniques
Bidard F, Saliba AE, Saias L, Degeorges A, Cremoux Pd, Viovy J, Vincent-Salomon A, Mathiot C, Pierga J, Gramont A (2009)
Bulletin du Cancer 96 (1): 73-86
2004
Nanotechnology serving biochips – The Toulouse example
Vieu C, Malaquin L, Thibault C, Saliba AE, Daran E, Dildan M, Carcenac F, Leberre V, Trevisiol E, François JM (2004)
Biofutur (250): 41-45

